Getting Started with Tilia Data Entry¶
Creating a New Tilia File¶
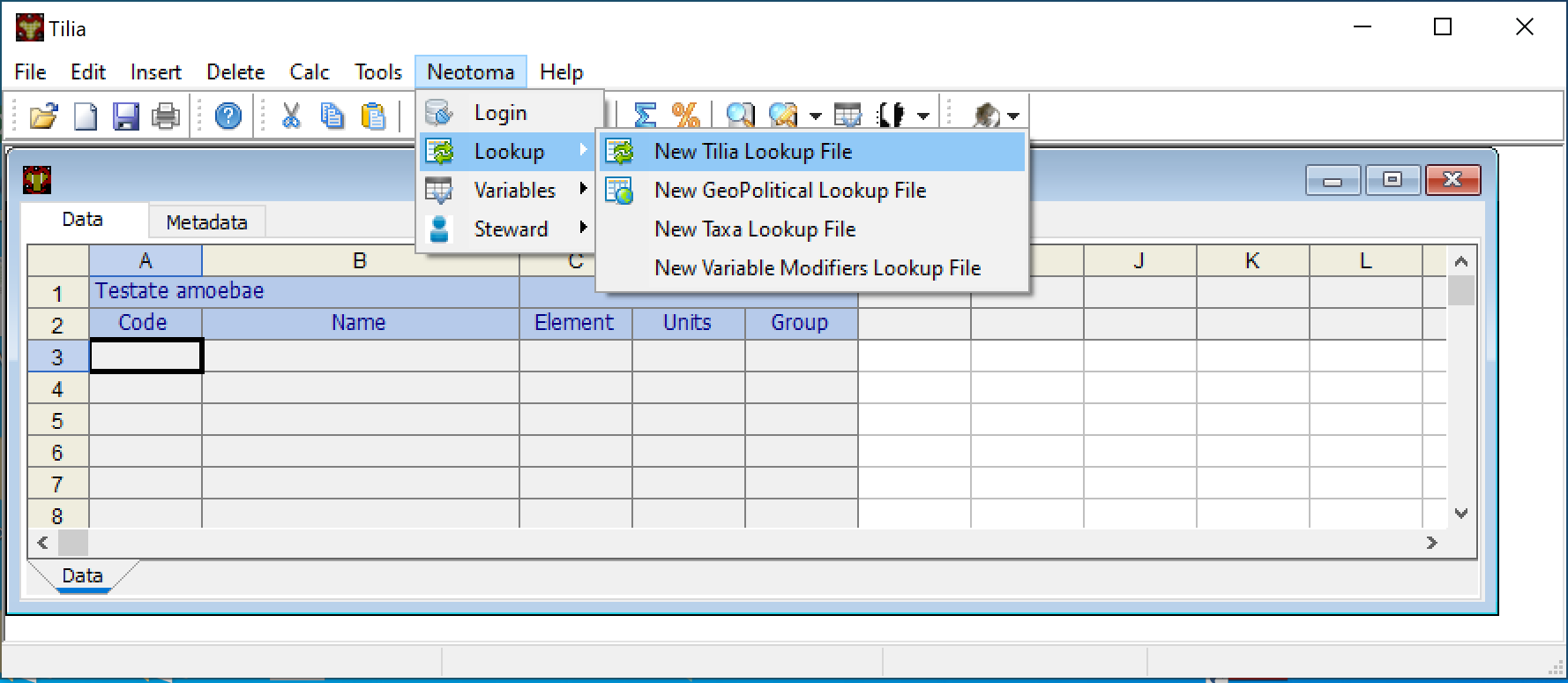
Open up the Tilia program and open a new file (File > New). A new window will open up, with two tabs at the top (Data & Metadata), several columns (Code, Name and Group) and a single header cell, by default Pollen.
The cell that contains the term Pollen can be modified to represent one of the other dataset types that Neotoma supports. To switch dataset types, click on the dropdown arrow to the right within the cell and select the appropriate data type.
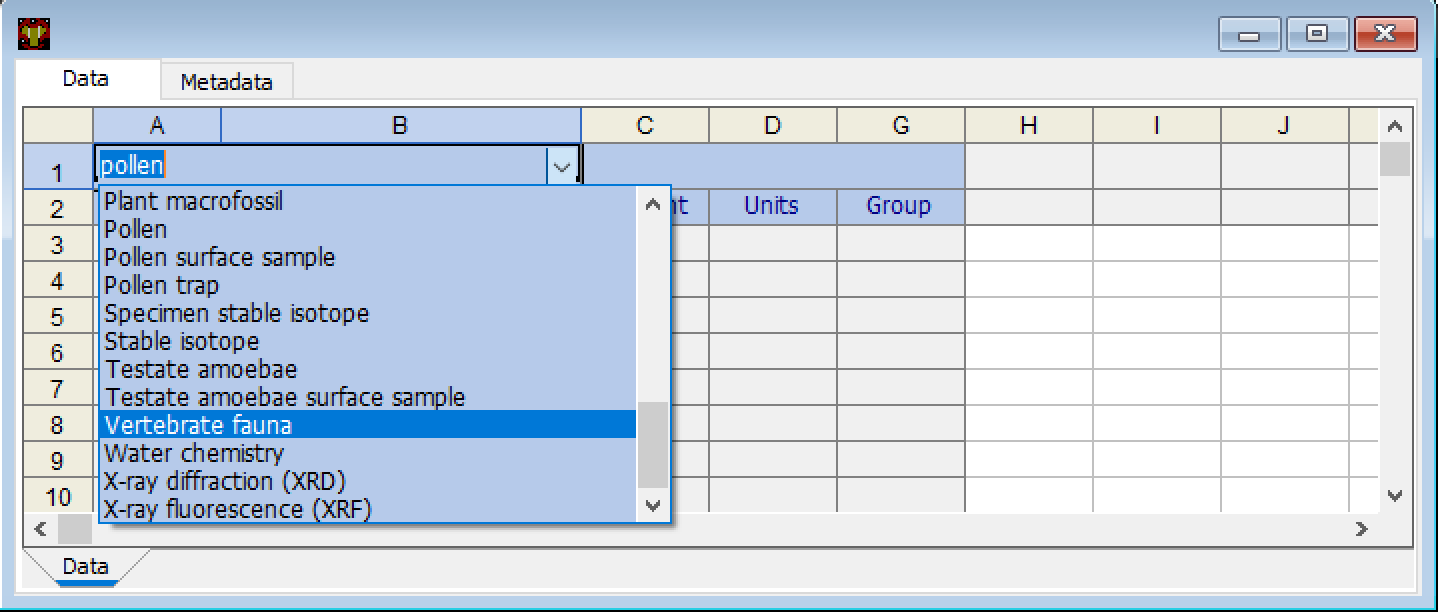
Changing data types for the subsequent rows of the spreadsheet.
Once you have selected the dataset type, it’s possible to then load the lookup files that Tilia uses to standardize taxonomy and other associated sample information.
Lookup Files¶
Tilia uses a series of lookup files that contain the structured variable entries available for different dataset types (taxa, elements, units, etc), depositional environments, geopolitical units, etc. Upon installation of Tilia, these lookup files will be downloaded to your local machine, and so every time you work with Tilia you need to ‘point’ Tilia to the appropriate lookup files for the datasets you are working with.
To load the lookup files, click Tools > Variable Lookup. The following window will appear:
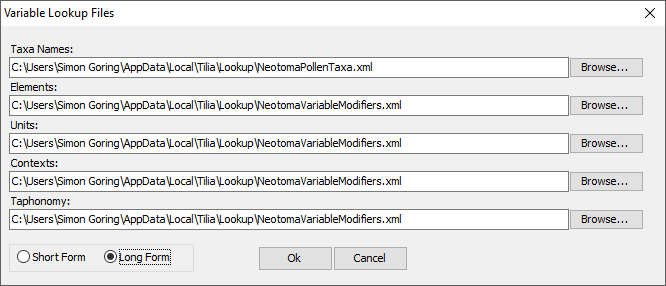
Then choose the tables that apply to the dataset type you are entering (you can use the Browse button if you don’t see what you need). In general, these will be auto-filled. This enables you to provide your own tables if needed, however, custom tables may make the upload & validation process more difficult for the Data Stewards. In the above figure, you can see the “Long Form” entry. The “Short Form” allows you to define only the Taxa Names and “Modifiers”. For the default lookup tables, there is no difference between the set of lookup files that are loaded with the “Short Form” vs “Long Form” entries.
Because of the way lookup tables are structured, it is often difficult to include two data types at the same time, since units or taphonomies specific to both proxies may not be available simultaneously.
A recommended practice is to periodically refresh the lookup files on your local machine to match those available through the database. This is particularly important in cases where new taxa are being added (for example, if a group is making a new push to enter data from a new region, and subsequently entering new taxa). To do this, click Neotoma > Lookup and then select the specific lookup file you would like to refresh.